JRM-UPO CSIC
Juan Ramón Martínez Morales
Juan Ramón Martínez-Morales (JRMM) group studies developmental genetics and tissue morphogenesis, with a particular focus on the early development of the vertebrate eye. Other topics such as the emergence and radiation of the vertebrate body plan and the coupling between mechanical forces and tissue morphogenesis are among the broad interests of the group.
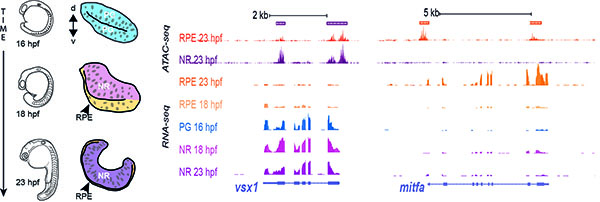
In the context of eye development, we explore how gene regulatory networks control tissue identity; how this translates in precise shape changes, as well as polarized contractility and cellular adhesion; and how cellular tensions are transmitted within the tissue to determine the final shape of the organ. The use of two far related teleost models (i.e. their lineages separated 200 mya) allows investigating the universality and relative contribution of the cellular and genetic mechanisms that control organ shape. In addition, due to the strategic position of teleost fish within the vertebrate evolutionary tree, our research often uncovers interesting evo-devo questions that are relevant to understand the evolutionary history of the vertebrate lineage. Therefore, questions such as, how new developmental mechanisms appear and how do they contribute to the emergence of vertebrate anatomical innovations, are among the research interests of the laboratory. During the last years, we have become increasingly aware that an important part of our developmental studies –particularly those exploring the divergence between the neural retina and retinal pigmented epithelium (RPE) genetic programs– are also relevant in the context of retinal diseases. This seems particularly the case for retinal degenerative pathologies, for which understanding the genetic program specifying the RPE is key to improve the efficiency of recently developed cell-based therapies.
In summary, although developmental genetics and tissue morphogenesis are the main interests in the group, we also pay attention to the other two vertices of the conceptual triangle connecting Evolution, Development, and Disease. From a methodological point of view, we use a broad set of techniques, including experimental embryology, live-imaging studies, fish genetics (CRISPR-Cas9, morpholinos, tilling), RNA-seq and scRNA-seq profiling, and epigenetic marks analysis (ChIP-seq, ATAC-seq, DamID-seq, and 4C-seq). This variety of methods enables a multidisciplinary approach to tackle questions often lying among developmental biology, disease modelling, and evolution.
Group Website:

